Details of experiments between Q96QZ7 and P60484
Experiment series 1 Protein A Protein: MAGI1 638-725 PDZ_3 his6-MBP-TEVsite-PDZ
Protein B Protein: PTEN 391-403 PBM EDQHTQITKV Biotin-ttds-PFDEDQHTQITKV
Measured dissociation constant (10-6 M):
Standard deviation of dissociation constant (10-6 M): 0
Average holdup BI: 0.58
Normalized holdup BI: 0
Normalized holdup BI standard deviation: 0
Immobilized partner concentration (10-6 M): 17
P value: 0
Experiment method: CALIP_holdup
Details of affinity fitting: lysozyme internal standard
Number of measurements: 1
Measured pK d : 4.98
Experiment series 2 Protein A Protein: MAGI1 986-1098 PDZ_5 his6-MBP-TEVsite-PDZ
Protein B Protein: PTEN 393-403 PBM EDQHTQITKV biotin-ttds-DEDQHTQITKV
Measured dissociation constant (10-6 M):
Standard deviation of dissociation constant (10-6 M): 0
Average holdup BI: 0.54
Normalized holdup BI: 0
Normalized holdup BI standard deviation: 0
Immobilized partner concentration (10-6 M): 36
P value: 0
Experiment method: CALIP_holdup
Details of affinity fitting: BSA and lysozyme internal standards + bacterial cell lysate + normal layout
Number of measurements: 1
Measured pK d : 4.53
Experiment series 3 Protein A Protein: MAGI1 638-725 PDZ_3 his6-MBP-TEVsite-PDZ
Protein B Protein: PTEN 393-403 PBM EDQHTQITKV biotin-ttds-DEDQHTQITKV
Measured dissociation constant (10-6 M):
Standard deviation of dissociation constant (10-6 M): 0
Average holdup BI: 0.52
Normalized holdup BI: 0
Normalized holdup BI standard deviation: 0
Immobilized partner concentration (10-6 M): 36
P value: 0
Experiment method: CALIP_holdup
Details of affinity fitting: BSA and lysozyme internal standards + bacterial cell lysate + normal layout
Number of measurements: 1
Measured pK d : 4.5
Experiment series 4 Protein A Protein: MAGI1 986-1098 PDZ_5 his6-MBP-TEVsite-PDZ
Protein B Protein: PTEN 391-403 PBM EDQHTQITKV Biotin-ttds-PFDEDQHTQITKV
Measured dissociation constant (10-6 M):
Standard deviation of dissociation constant (10-6 M): 0
Average holdup BI: 0.27
Normalized holdup BI: 0
Normalized holdup BI standard deviation: 0
Immobilized partner concentration (10-6 M): 17
P value: 0
Experiment method: CALIP_holdup
Details of affinity fitting: lysozyme internal standard
Number of measurements: 1
Measured pK d : 4.38
Experiment series 5 Protein A Protein: MAGI1 1-106 PDZ_1 his6-MBP-TEVsite-PDZ
Protein B Protein: PTEN 391-403 PBM EDQHTQITKV Biotin-ttds-PFDEDQHTQITKV
Measured dissociation constant (10-6 M):
Standard deviation of dissociation constant (10-6 M): 0
Average holdup BI: -0.03
Normalized holdup BI: 0
Normalized holdup BI standard deviation: 0
Immobilized partner concentration (10-6 M): 17
P value: 0
Experiment method: CALIP_holdup
Details of affinity fitting: lysozyme internal standard
Number of measurements: 1
The affinity was below the detection threshold of the assay.
Experiment series 6 Protein A Protein: MAGI1 1-106 PDZ_1 his6-MBP-TEVsite-PDZ
Protein B Protein: PTEN 393-403 PBM EDQHTQITKV biotin-ttds-DEDQHTQITKV
Measured dissociation constant (10-6 M):
Standard deviation of dissociation constant (10-6 M): 0
Average holdup BI: 0
Normalized holdup BI: 0
Normalized holdup BI standard deviation: 0
Immobilized partner concentration (10-6 M): 36
P value: 0
Experiment method: CALIP_holdup
Details of affinity fitting: BSA and lysozyme internal standards + bacterial cell lysate + normal layout
Number of measurements: 1
The affinity was below the detection threshold of the assay.
Experiment series 7 Protein A Protein: MAGI1 1-106 PDZ_1 his6-MBP-TEVsite-PDZ
Protein B Protein: PTEN 393-403 PBM EDQHTQITKV biotin-ttds-DEDQHTQITKV
Measured dissociation constant (10-6 M):
Standard deviation of dissociation constant (10-6 M): 0
Average holdup BI: 0.05
Normalized holdup BI: 0.05
Normalized holdup BI standard deviation: 0
Immobilized partner concentration (10-6 M): 19
P value: 0
Experiment method: DAPF_holdup
Details of affinity fitting: fluorescein + mCherry internal standards + normalization based on CALIP_holdup data + reverse layout
Number of measurements: 1
The affinity was below the detection threshold of the assay.
Experiment series 8 Protein A Protein: MAGI1 1-106 PDZ_1 his6-MBP-TEVsite-PDZ
Protein B Protein: PTEN 394-403 PBM EDQHTQITKV Biotin-ado-ado-EDQHTQITKV
Measured dissociation constant (10-6 M):
Standard deviation of dissociation constant (10-6 M): 0
Average holdup BI: 0.01
Normalized holdup BI: 0.01
Normalized holdup BI standard deviation: 0
Immobilized partner concentration (10-6 M): 18
P value: 0
Experiment method: DAPF_holdup
Details of affinity fitting: fluorescein + mCherry internal standards + normalization based on CALIP_holdup data + reverse layout
Number of measurements: 1
The affinity was below the detection threshold of the assay.
Experimental Details Crude peptide synthesis product. The peptide concentration in the affinity conversion step may differ from reality. Not reported internal standards of holdup layout #6
Experiment series 9 Protein A Protein: MAGI1 1146-1238 PDZ_6 his6-MBP-TEVsite-PDZ
Protein B Protein: PTEN 391-403 PBM EDQHTQITKV Biotin-ttds-PFDEDQHTQITKV
Measured dissociation constant (10-6 M):
Standard deviation of dissociation constant (10-6 M): 0
Average holdup BI: 0.07
Normalized holdup BI: 0
Normalized holdup BI standard deviation: 0
Immobilized partner concentration (10-6 M): 17
P value: 0
Experiment method: CALIP_holdup
Details of affinity fitting: lysozyme internal standard
Number of measurements: 1
The affinity was below the detection threshold of the assay.
Experiment series 10 Protein A Protein: MAGI1 1146-1238 PDZ_6 his6-MBP-TEVsite-PDZ
Protein B Protein: PTEN 393-403 PBM EDQHTQITKV biotin-ttds-DEDQHTQITKV
Measured dissociation constant (10-6 M):
Standard deviation of dissociation constant (10-6 M): 0
Average holdup BI: -0.04
Normalized holdup BI: 0
Normalized holdup BI standard deviation: 0
Immobilized partner concentration (10-6 M): 36
P value: 0
Experiment method: CALIP_holdup
Details of affinity fitting: BSA and lysozyme internal standards + bacterial cell lysate + normal layout
Number of measurements: 1
The affinity was below the detection threshold of the assay.
Experiment series 11 Protein A Protein: MAGI1 456-580 PDZ_2 his6-MBP-TEVsite-PDZ
Protein B Protein: PTEN 391-403 PBM EDQHTQITKV Biotin-ttds-PFDEDQHTQITKV
Measured dissociation constant (10-6 M):
Standard deviation of dissociation constant (10-6 M): 0
Average holdup BI: -0.02
Normalized holdup BI: 0
Normalized holdup BI standard deviation: 0
Immobilized partner concentration (10-6 M): 17
P value: 0
Experiment method: CALIP_holdup
Details of affinity fitting: lysozyme internal standard
Number of measurements: 1
The affinity was below the detection threshold of the assay.
Experiment series 12 Protein A Protein: MAGI1 456-580 PDZ_2 his6-MBP-TEVsite-PDZ
Protein B Protein: PTEN 393-403 PBM EDQHTQITKV biotin-ttds-DEDQHTQITKV
Measured dissociation constant (10-6 M):
Standard deviation of dissociation constant (10-6 M): 0
Average holdup BI: 0.02
Normalized holdup BI: 0.03
Normalized holdup BI standard deviation: 0
Immobilized partner concentration (10-6 M): 19
P value: 0
Experiment method: DAPF_holdup
Details of affinity fitting: fluorescein + mCherry internal standards + normalization based on CALIP_holdup data + reverse layout
Number of measurements: 1
The affinity was below the detection threshold of the assay.
Experiment series 13 Protein A Protein: MAGI1 456-580 PDZ_2 his6-MBP-TEVsite-PDZ
Protein B Protein: PTEN 393-403 PBM EDQHTQITKV biotin-ttds-DEDQHTQITKV
Measured dissociation constant (10-6 M):
Standard deviation of dissociation constant (10-6 M): 0
Average holdup BI: 0.08
Normalized holdup BI: 0
Normalized holdup BI standard deviation: 0
Immobilized partner concentration (10-6 M): 36
P value: 0
Experiment method: CALIP_holdup
Details of affinity fitting: BSA and lysozyme internal standards + bacterial cell lysate + normal layout
Number of measurements: 1
The affinity was below the detection threshold of the assay.
Experiment series 14 Protein A Protein: MAGI1 456-580 PDZ_2 his6-MBP-TEVsite-PDZ
Protein B Protein: PTEN 393-403 PBM EDQHTQITKV biotin-ttds-DEDQHTQITKV
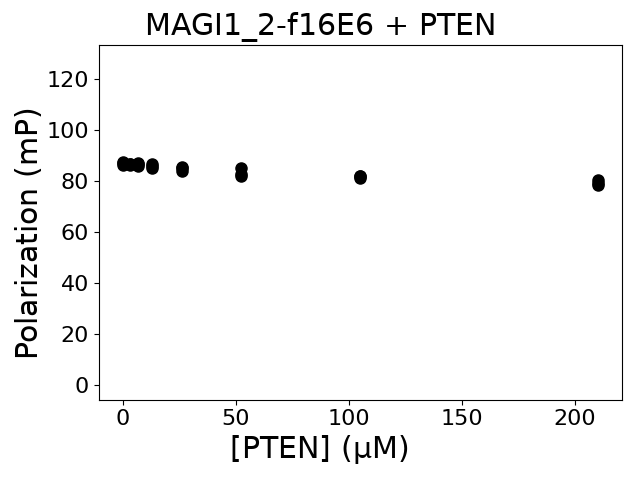
Measured dissociation constant (10-6 M): 0
Standard deviation of dissociation constant (10-6 M): 0
Average holdup BI: 0
Normalized holdup BI: 0
Normalized holdup BI standard deviation: 0
Immobilized partner concentration (10-6 M): 0
P value: 0
Experiment method: competitive FP
Details of affinity fitting: Competitive binding equation in ProFit
Number of measurements: 3
The affinity was below the detection threshold of the assay.
Experiment series 15 Protein A Protein: MAGI1 456-580 PDZ_2 his6-MBP-TEVsite-PDZ
Protein B Protein: PTEN 394-403 PBM EDQHTQITKV Biotin-ado-ado-EDQHTQITKV
Measured dissociation constant (10-6 M):
Standard deviation of dissociation constant (10-6 M): 0
Average holdup BI: 0.01
Normalized holdup BI: 0.01
Normalized holdup BI standard deviation: 0
Immobilized partner concentration (10-6 M): 18
P value: 0
Experiment method: DAPF_holdup
Details of affinity fitting: fluorescein + mCherry internal standards + normalization based on CALIP_holdup data + reverse layout
Number of measurements: 1
The affinity was below the detection threshold of the assay.
Experimental Details Crude peptide synthesis product. The peptide concentration in the affinity conversion step may differ from reality. Not reported internal standards of holdup layout #6
Experiment series 16 Protein A Protein: MAGI1 836-926 PDZ_4 his6-MBP-TEVsite-PDZ
Protein B Protein: PTEN 391-403 PBM EDQHTQITKV Biotin-ttds-PFDEDQHTQITKV
Measured dissociation constant (10-6 M):
Standard deviation of dissociation constant (10-6 M): 0
Average holdup BI: 0.09
Normalized holdup BI: 0
Normalized holdup BI standard deviation: 0
Immobilized partner concentration (10-6 M): 17
P value: 0
Experiment method: CALIP_holdup
Details of affinity fitting: lysozyme internal standard
Number of measurements: 1
The affinity was below the detection threshold of the assay.
Experiment series 17 Protein A Protein: MAGI1 638-725 PDZ_3 his6-MBP-TEVsite-PDZ
Protein B Protein: PTEN 393-403 PBM (ALY)402 EDQHTQITKV biotin-ttds-DEDQHTQITKacV
Measured dissociation constant (10-6 M):
Standard deviation of dissociation constant (10-6 M): 0
Average holdup BI: 0.72
Normalized holdup BI: 0
Normalized holdup BI standard deviation: 0
Immobilized partner concentration (10-6 M): 29
P value: 0
Experiment method: CALIP_holdup
Details of affinity fitting: BSA and lysozyme internal standards + bacterial cell lysate + normal layout
Number of measurements: 1
Measured pK d : 4.98
Experiment series 18 Protein A Protein: MAGI1 986-1098 PDZ_5 his6-MBP-TEVsite-PDZ
Protein B Protein: PTEN 393-403 PBM (ALY)402 EDQHTQITKV biotin-ttds-DEDQHTQITKacV
Measured dissociation constant (10-6 M):
Standard deviation of dissociation constant (10-6 M): 0
Average holdup BI: 0.65
Normalized holdup BI: 0
Normalized holdup BI standard deviation: 0
Immobilized partner concentration (10-6 M): 29
P value: 0
Experiment method: CALIP_holdup
Details of affinity fitting: BSA and lysozyme internal standards + bacterial cell lysate + normal layout
Number of measurements: 1
Measured pK d : 4.84
Experiment series 19 Protein A Protein: MAGI1 456-580 PDZ_2 his6-MBP-TEVsite-PDZ
Protein B Protein: PTEN 393-403 PBM (ALY)402 EDQHTQITKV biotin-ttds-DEDQHTQITKacV
Measured dissociation constant (10-6 M):
Standard deviation of dissociation constant (10-6 M): 0
Average holdup BI: 0.07
Normalized holdup BI: 0.08
Normalized holdup BI standard deviation: 0
Immobilized partner concentration (10-6 M): 17
P value: 0
Experiment method: DAPF_holdup
Details of affinity fitting: fluorescein + mCherry internal standards + normalization based on CALIP_holdup data + reverse layout
Number of measurements: 1
Measured pK d : 3.74
Experimental Details not reported internal standards of holdup layout #5
Experiment series 20 Protein A Protein: MAGI1 456-580 PDZ_2 his6-MBP-TEVsite-PDZ
Protein B Protein: PTEN 393-403 PBM (ALY)402 EDQHTQITKV biotin-ttds-DEDQHTQITKacV
Measured dissociation constant (10-6 M):
Standard deviation of dissociation constant (10-6 M): 0
Average holdup BI: 0.07
Normalized holdup BI: 0.08
Normalized holdup BI standard deviation: 0
Immobilized partner concentration (10-6 M): 17
P value: 0
Experiment method: DAPF_holdup
Details of affinity fitting: fluorescein + mCherry internal standards + normalization based on CALIP_holdup data + reverse layout
Number of measurements: 1
Measured pK d : 3.69
Experiment series 21 Protein A Protein: MAGI1 456-580 PDZ_2 his6-MBP-TEVsite-PDZ
Protein B Protein: PTEN 393-403 PBM (ALY)402 EDQHTQITKV biotin-ttds-DEDQHTQITKacV
Measured dissociation constant (10-6 M):
Standard deviation of dissociation constant (10-6 M): 0
Average holdup BI: 0.1
Normalized holdup BI: 0
Normalized holdup BI standard deviation: 0
Immobilized partner concentration (10-6 M): 29
P value: 0
Experiment method: CALIP_holdup
Details of affinity fitting: BSA and lysozyme internal standards + bacterial cell lysate + normal layout
Number of measurements: 1
Measured pK d : 3.59
Experiment series 22 Protein A Protein: MAGI1 1-106 PDZ_1 his6-MBP-TEVsite-PDZ
Protein B Protein: PTEN 393-403 PBM (ALY)402 EDQHTQITKV biotin-ttds-DEDQHTQITKacV
Measured dissociation constant (10-6 M):
Standard deviation of dissociation constant (10-6 M): 0
Average holdup BI: 0.01
Normalized holdup BI: 0.01
Normalized holdup BI standard deviation: 0
Immobilized partner concentration (10-6 M): 17
P value: 0
Experiment method: DAPF_holdup
Details of affinity fitting: fluorescein + mCherry internal standards + normalization based on CALIP_holdup data + reverse layout
Number of measurements: 1
The affinity was below the detection threshold of the assay.
Experiment series 23 Protein A Protein: MAGI1 1-106 PDZ_1 his6-MBP-TEVsite-PDZ
Protein B Protein: PTEN 393-403 PBM (ALY)402 EDQHTQITKV biotin-ttds-DEDQHTQITKacV
Measured dissociation constant (10-6 M):
Standard deviation of dissociation constant (10-6 M): 0
Average holdup BI: 0.02
Normalized holdup BI: 0.02
Normalized holdup BI standard deviation: 0
Immobilized partner concentration (10-6 M): 17
P value: 0
Experiment method: DAPF_holdup
Details of affinity fitting: fluorescein + mCherry internal standards + normalization based on CALIP_holdup data + reverse layout
Number of measurements: 1
The affinity was below the detection threshold of the assay.
Experimental Details not reported internal standards of holdup layout #5
Experiment series 24 Protein A Protein: MAGI1 1-106 PDZ_1 his6-MBP-TEVsite-PDZ
Protein B Protein: PTEN 393-403 PBM (ALY)402 EDQHTQITKV biotin-ttds-DEDQHTQITKacV
Measured dissociation constant (10-6 M):
Standard deviation of dissociation constant (10-6 M): 0
Average holdup BI: 0.03
Normalized holdup BI: 0
Normalized holdup BI standard deviation: 0
Immobilized partner concentration (10-6 M): 29
P value: 0
Experiment method: CALIP_holdup
Details of affinity fitting: BSA and lysozyme internal standards + bacterial cell lysate + normal layout
Number of measurements: 1
The affinity was below the detection threshold of the assay.
Experiment series 25 Protein A Protein: MAGI1 1-106 PDZ_1 his6-MBP-TEVsite-PDZ
Protein B Protein: PTEN 393-403 PBM (ALY)402 EDQHTQITKV biotin-ttds-DEDQHTQITKacV
Measured dissociation constant (10-6 M):
Standard deviation of dissociation constant (10-6 M): 0
Average holdup BI: 0.04
Normalized holdup BI: 0.04
Normalized holdup BI standard deviation: 0
Immobilized partner concentration (10-6 M): 18
P value: 0
Experiment method: DAPF_holdup
Details of affinity fitting: fluorescein + mCherry internal standards + normalization based on CALIP_holdup data + reverse layout
Number of measurements: 1
The affinity was below the detection threshold of the assay.
Experimental Details not reported internal standards of holdup layout #6
Experiment series 26 Protein A Protein: MAGI1 1146-1238 PDZ_6 his6-MBP-TEVsite-PDZ
Protein B Protein: PTEN 393-403 PBM (ALY)402 EDQHTQITKV biotin-ttds-DEDQHTQITKacV
Measured dissociation constant (10-6 M):
Standard deviation of dissociation constant (10-6 M): 0
Average holdup BI: 0.1
Normalized holdup BI: 0
Normalized holdup BI standard deviation: 0
Immobilized partner concentration (10-6 M): 29
P value: 0
Experiment method: CALIP_holdup
Details of affinity fitting: BSA and lysozyme internal standards + bacterial cell lysate + normal layout
Number of measurements: 1
The affinity was below the detection threshold of the assay.
Experiment series 27 Protein A Protein: MAGI1 456-580 PDZ_2 his6-MBP-TEVsite-PDZ
Protein B Protein: PTEN 393-403 PBM (ALY)402 EDQHTQITKV biotin-ttds-DEDQHTQITKacV
Measured dissociation constant (10-6 M):
Standard deviation of dissociation constant (10-6 M): 0
Average holdup BI: 0.04
Normalized holdup BI: 0.05
Normalized holdup BI standard deviation: 0
Immobilized partner concentration (10-6 M): 18
P value: 0
Experiment method: DAPF_holdup
Details of affinity fitting: fluorescein + mCherry internal standards + normalization based on CALIP_holdup data + reverse layout
Number of measurements: 1
The affinity was below the detection threshold of the assay.
Experimental Details not reported internal standards of holdup layout #6
Experiment series 28 Protein A Protein: MAGI1 456-580 PDZ_2 his6-MBP-TEVsite-PDZ
Protein B Protein: PTEN 393-403 PBM (ALY)402 EDQHTQITKV biotin-ttds-DEDQHTQITKacV
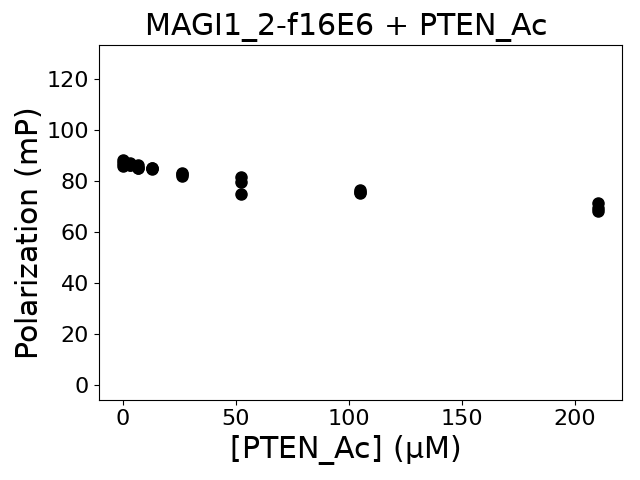
Measured dissociation constant (10-6 M): 0
Standard deviation of dissociation constant (10-6 M): 0
Average holdup BI: 0
Normalized holdup BI: 0
Normalized holdup BI standard deviation: 0
Immobilized partner concentration (10-6 M): 0
P value: 0
Experiment method: competitive FP
Details of affinity fitting: Competitive binding equation in ProFit
Number of measurements: 3
The affinity was below the detection threshold of the assay.
Please cite ProfAff! (Gogl, G. et al., Nature Communications 2022) research team of Gergo Gogl .