Details of experiments between Q7Z628 and Q96L92
Note that the results of all experiments are listed, regardless of the modification states of the fragments.
Crude peptide synthesis product. The peptide concentration in the affinity conversion step may differ from reality. Not reported internal standards of holdup layout #6
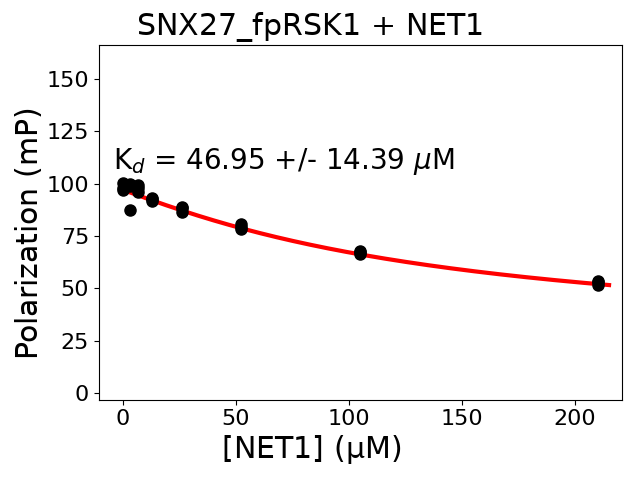
not reported internal standards of holdup layout #5
not reported internal standards of holdup layout #5