Details of experiments between Q7Z460 and Q04917
Note that the results of all experiments are listed, regardless of the modification states of the fragments.
-log10(P Two-sided unpaired T-test (4 vs 5))
-log10(P Two-sided unpaired T-test (4 vs 5))
-log10(P Two-sided unpaired T-test (4 vs 5))
-log10(P Two-sided unpaired T-test (4 vs 5))
-log10(P Two-sided unpaired T-test (4 vs 5))
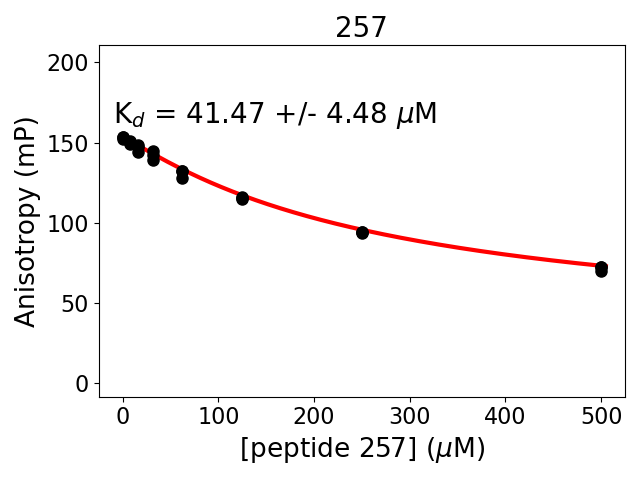
-log10(P Two-sided unpaired T-test (4 vs 5))
-log10(P Two-sided unpaired T-test (4 vs 5))
-log10(P Two-sided unpaired T-test (4 vs 5))
-log10(P Two-sided unpaired T-test (4 vs 5))
-log10(P Two-sided unpaired T-test (4 vs 5))