Average holdup BI: 0.04
Holdup BI standard deviation: 0.03
Normalized holdup BI: 0.04
Normalized holdup BI standard deviation: 0.04
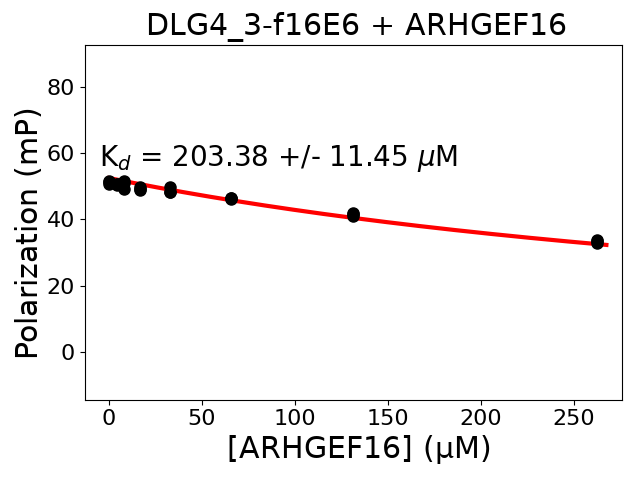
Immobilized partner concentration (10-6M): 17
Experiment method: DAPF_holdup
Details of affinity fitting: fluorescein + mCherry internal standards + normalization based on CALIP_holdup data + reverse layout
Number of measurements: 2