Details of experiments between Q15311 and Q96L92
Note that the results of all experiments are listed, regardless of the modification states of the fragments.
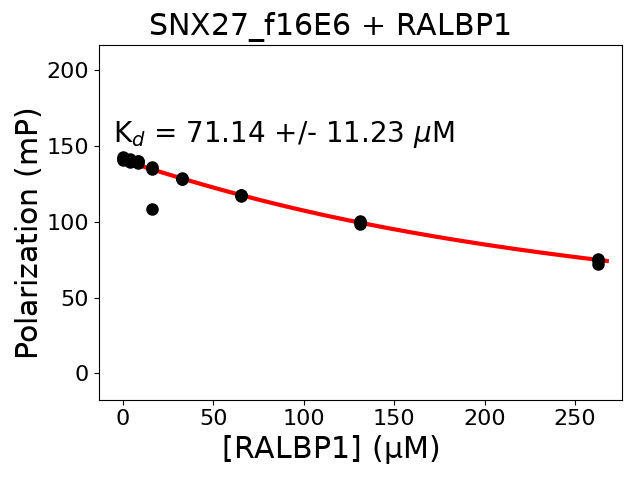
not reported internal standards of holdup layout #6
-log10(P Two-sided unpaired T-test (6 vs 3))